DrugFormDB: A GPT-4 & PubMedBERT Drug dataset
How I classified 3.8K medications into their respective forms to improve MARMAR's user flow and personalization result.
As you might already know, one of the seven questions we ask when you use MARMAR is, “What medication are you currently taking?”. So while I was ideating how we can give our users an even more personalized report, I started to think of additional relevant questions to ask. After a bit of research, I figured out that asking a follow-up question about the dosage of their medication is also paramount.
The concept is simple: the moment a user enters their medication name, our system already knows whether it's a tablet, injection, cream, or one of nine other standardized forms. This means we can immediately ask the right dosage question—"How many milligrams (mg) per dose?" for tablets or "How many international units (IU) per injection?”—without first asking users to specify the form themselves, you know just part of the 'little things that makes the flow smoother
This article outlines my process for building this dataset, leveraging GPT-4 for data generation and PubMedBERT's domain-specific knowledge for validation. I'll share my methodology, the challenges I encountered, and the solutions I developed along the way.
The Data Challenge: Starting With Just Drug Names
The initial dataset comprised +3k drug names from regulatory databases listed in a JSON file. The challenge was in the complexity of modern pharmaceuticals. A single drug might come in multiple forms, and new delivery methods are constantly emerging. I wanted a system that could handle this complexity while remaining accurate and maintainable.
Solution: A Two-Stage Classification System
Stage 1: GPT-4 Classification
I chose GPT-4 as our primary classifier for several reasons:
Contextual Understanding: GPT-4 can interpret drug names in their pharmaceutical context, understanding that "Amoxicillin" might come in multiple forms while "Insulin" is primarily an injection, subcutaneous .
Structured Output: I engineered the prompt to ensure standardized outputs, and limited the forms into 12 differnt standardized forms, if any of the medciations falls outside of these listed forms we should get an unknown.
VALID_FORMS = [ "Tablet", "Capsule", "Oral Solution", "Oral Suspension/Syrup", "Injection/Infusion", "Cream", "Ointment", "Gel", "Patch", "Inhaler/Nasal Spray", "Eye/Ear Drops", "Suppository" ]Handling of Uncertainty: Rather than guessing, I made sure GPT-4 marks unclear cases as "Unknown".
Technical Implementation Details
Our classification script (classify_drugs.py) incorporated several critical features:
Batch Processing with Smart Recovery:
BATCH_SIZE = 100 # Optimized for API efficiency for i in range(0, len(drugs_to_process), BATCH_SIZE): batch = drugs_to_process[i:i + BATCH_SIZE] # Process and save after each drug for resilienceContinuous Progress Tracking:
Real-time progress updates
Automatic saving after each classification
Ability to resume from interruptions
So this means by the end of it all I had a JSON file of 3,856 drugs with their respective forms.
The prompt :
You are a pharmaceutical expert with deep knowledge of drug formulations and delivery methods. Consider the drug's active ingredients, therapeutic use, and typical administration routes. Then I constrained its output to exactly 12 standardized forms: Tablet", "Capsule", "Oral Solution", "Oral Suspension/Syrup", "Injection/Infusion", "Cream", "Ointment", "Gel", "Patch", "Inhaler/Nasal Spray", "Eye/Ear Drops", "Suppository"
The results were fascinating. Take amoxicillin, for example: GPT-4 correctly identified it comes in "Tablet, Capsule, Oral Suspension/Syrup". It understood that antibiotics often need multiple forms for different patient needs; the model even maintained consistency with real-world pharmaceutical practices. But it begs the question: how was it validated, and how did I know, right? Flow with me.
Semantic Similarity Validation
To validate GPT-4's classifications, I implemented a validation approach using PubMedBERT. PubMedBERT is not a generative model like GPT-4. It's a BERT-based transformer model specifically fine-tuned on biomedical literature, and its key characteristics are:
Pre-trained on: PubMed abstracts and full-text articles
Architecture: Based on BERT (Bidirectional Encoder Representations from Transformers)
Specialization: Medical and pharmaceutical domain knowledge
Primary function: Semantic understanding of medical text
To be more specific, I used a variant of PubMed called “pritamdeka/S-PubMedBert-MS-MARCO,” which is further optimized for semantic similarity tasks, and its fine-tuning on medical literature and the MS-MARCO dataset which makes it particularly suited for understanding medical/health contexts.
The validation process is interesting because of how I structured the queries to PubMedBERT. For each drug that GPT-4 has classified (+3000 of them), I created a natural language statement and asked PubMedBERT to understand it semantically.
Here's the detailed process:
test_statement = f"{drug_name} is administered as a {form}"
test_embedding = model.encode(test_statement)To explain this further, what I did was to take a drug name:
drug_name = "Amoxicillin"
gpt4_form = "Tablet"Then I created a test statements:
# For GPT-4's suggested form
"Amoxicillin is administered as a Tablet"
# Behind the scenes, I also test against all other (12) forms:
"Amoxicillin is administered as a Capsule"
"Amoxicillin is administered as a Oral Solution"
# ... and so on for other formsWhat PubMedBERT Does behing the scene:
Takes each statement
Converts it into a 768-dimensional vector (embedding)
This vector captures the semantic meaning of the drug-form relationship
The vector includes understanding from medical literature about:
The drug's typical uses
Common administration methods
Medical context
Pharmaceutical properties
Comparison Process:
# For each drug, we end up with multiple embeddings:
amoxicillin_tablet = model.encode("Amoxicillin is administered as a Tablet")
amoxicillin_capsule = model.encode("Amoxicillin is administered as a Capsule")
# ... etc.
# These are compared against our reference embeddings:
tablet_reference = reference_embeddings["Tablet"]
capsule_reference = reference_embeddings["Capsule"]
# ... etc.Real Example with Scores:
Drug: "Amoxicillin"
GPT-4's classification: "Tablet"
Generated statements and their similarity scores:
- "Amoxicillin is administered as a Tablet"
→ Similarity: 0.95 (High confidence, matches medical knowledge)
- "Amoxicillin is administered as a Capsule"
→ Similarity: 0.87 (Also high, as Amoxicillin does come in capsule form)
- "Amoxicillin is administered as a Injection/Infusion"
→ Similarity: 0.32 (Low, as this isn't a common form for Amoxicillin)Why This Works: PubMedBERT has been trained on millions of medical documents, as stated earlier, so it understands that:
Amoxicillin is commonly given orally
Tablets and capsules are typical forms
It's rarely given as an injection, hence the low score for the injection comparison.
As you will see, the final validation result:
Final Validation Results:
{
"Tablet": 0.95, # Very confident
"Capsule": 0.87, # Also quite confident
"Oral Solution": 0.82, # Possible form
"Injection": 0.32, # Unlikely form
# ... other forms
}This tells us that not only was GPT-4's "Tablet" classification correct, but also that "Capsule" and "Oral Solution" are reasonable alternatives, while "injection" is not—which matches real-world pharmaceutical knowledge.
The similarity score taken is basically calculated using the Cosine Similarity
similarity = np.dot(test_embedding, ref_embedding) / (
np.linalg.norm(test_embedding) * np.linalg.norm(ref_embedding)
)What we are essentially doing is
Taking the two vectors (test and reference embeddings)
Calculating their dot product
Normalizing it by their magnitudes
This produces a score between -1 and 1 (in practice, usually 0 to 1)
You can read more about cosine similarity here. So essentially this is what I did for each drug name that I had—generate its corresponding form using GPT-4, then proceed to validate it using PubMedBERT.
So essentially when we validate a drug's form, we get two key pieces of information:
1. Similarity Score: How confident PubMedBERT is about GPT-4's suggestion
2. Best Match: Which form got the highest similarity score
For example, with Amoxicillin:
When GPT-4 suggests "Tablet":
{
"drug": "Amoxicillin",
"gpt4_form": "Tablet",
"similarity_score": 0.95, # Confidence in "Tablet"
"best_match": "Tablet", # Form with highest score
"best_match_score": 0.95, # Same as similarity_score because they agree
"agrees_with_gpt4": True # They match!
}
But for a disagreement case (like Biotin):
{
"drug": "Biotin",
"gpt4_form": "Tablet",
"similarity_score": 0.87, # Confidence in "Tablet"
"best_match": "Capsule", # PubMedBERT thinks it's a capsule
"best_match_score": 0.92, # Higher than similarity_score
"agrees_with_gpt4": False # They disagree!
}This validation helps us in two ways:
1. Confirms when GPT-4 is right (high similarity score AND agreement)
2. Catches potential errors (when PubMedBERT finds a better match)
The Boolean Agreement
The Boolean agreement, which is simply a direct comparison—True if GPT-4's form matches PubMedBERT's best match and False if they differ. See full example below:
Drug: "Amoxicillin"
GPT-4 classification: "Tablet"
1. Create test statement:
"Amoxicillin is administered as a tablet"
2. Generate embeddings:
- Convert to 768-dimensional vector
- Compare with all reference embeddings
3. Calculate similarities:
Tablet: 0.95
Capsule: 0.87
Oral Solution: 0.82
...
4. Results:
similarity_score: 0.95 (for "Tablet")
best_match: "Tablet"
best_match_score: 0.95
agrees_with_gpt4: TrueThe validation process is implemented in drug_validator.py - and you can get this on my GitHub.
Overall Visualizations:
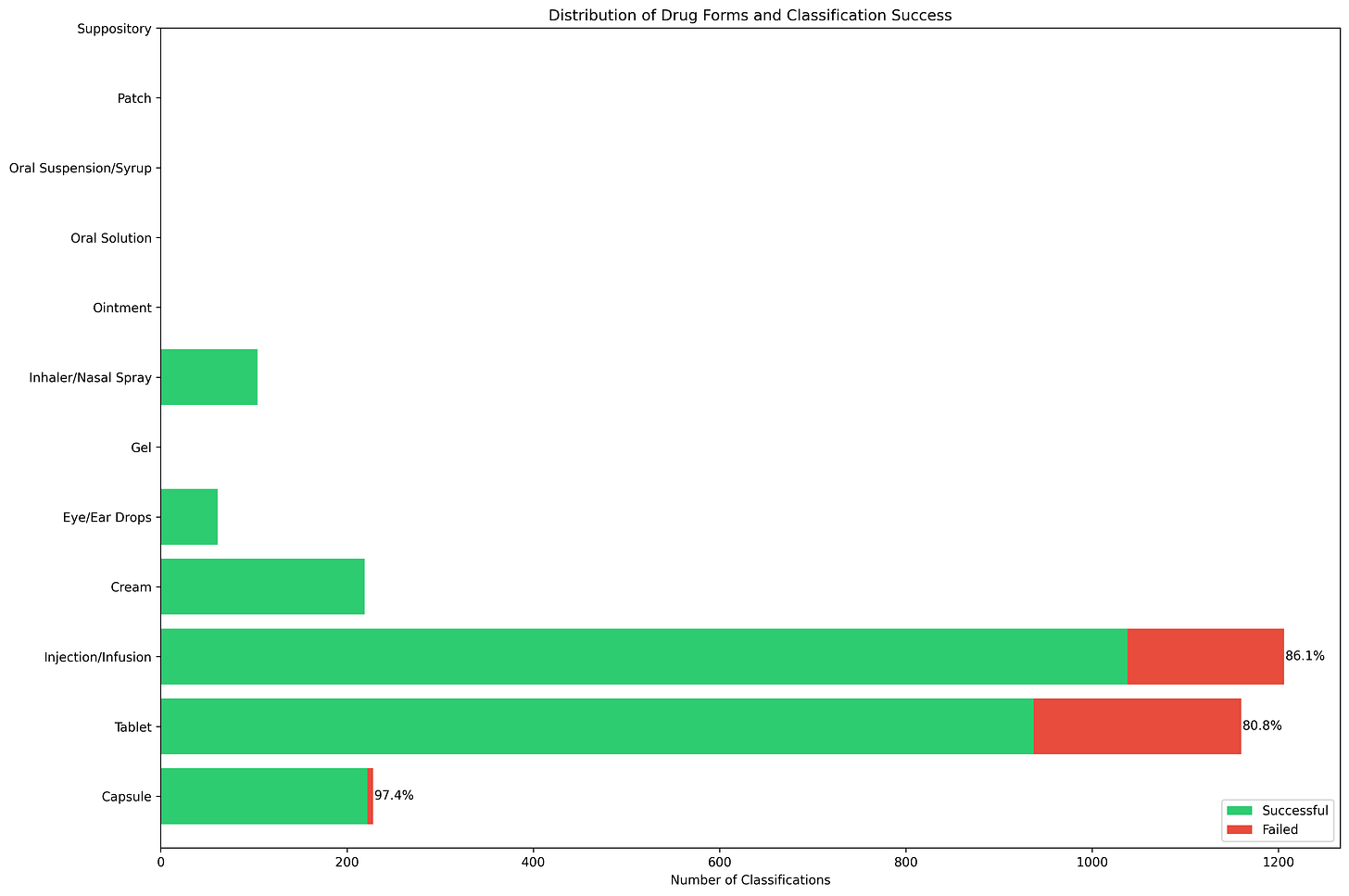
This shows the distribution of different drug forms dataset, with tablets and injections/infusions being the most numerous categories, followed by capsules and creams, as you can see, and oral solutions, patches, and suppositories appear less frequently. The percentages (86.1%, 80.8%, 97.4%) shows the success rates for the most common forms. Capsules have the highest success rate at 97.4%, while tablets and injections show moderate success rates around 80-86%.
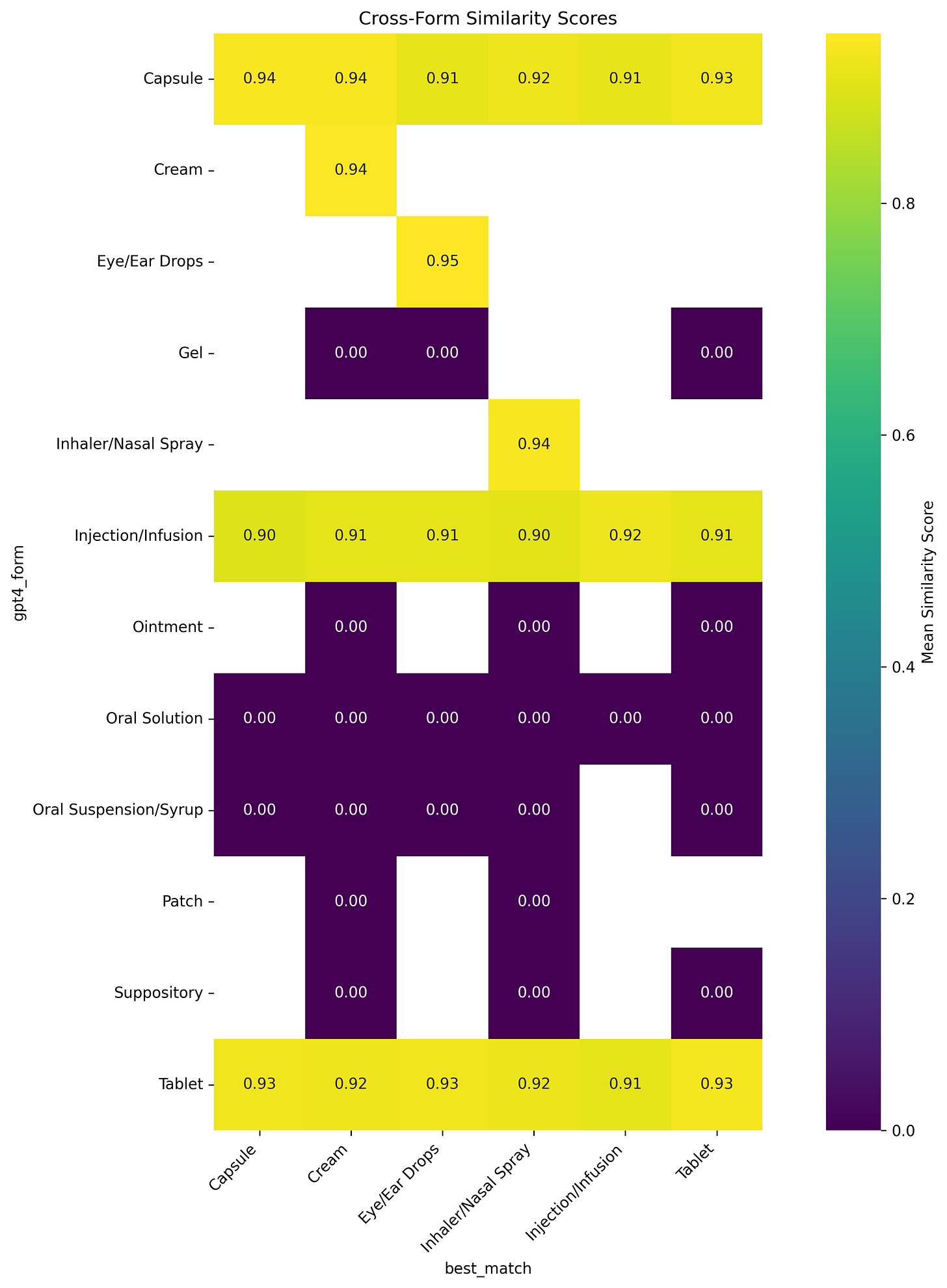
The similarity heatmap shows us a window into how it perceives relationships between different drug forms. It shows the distinct clusters of related forms, with particularly strong associations among topical preparations (creams, gels, and ointments) and oral formulations (tablets, capsules, and solutions).
For instance, the high similarity scores between creams and gels (typically 0.85-0.90) explain many of our classification challenges with topical medications. These patterns reflect not just classification difficulties but also the genuine pharmaceutical relationships between different forms, where similar administration routes often share physical and chemical properties.
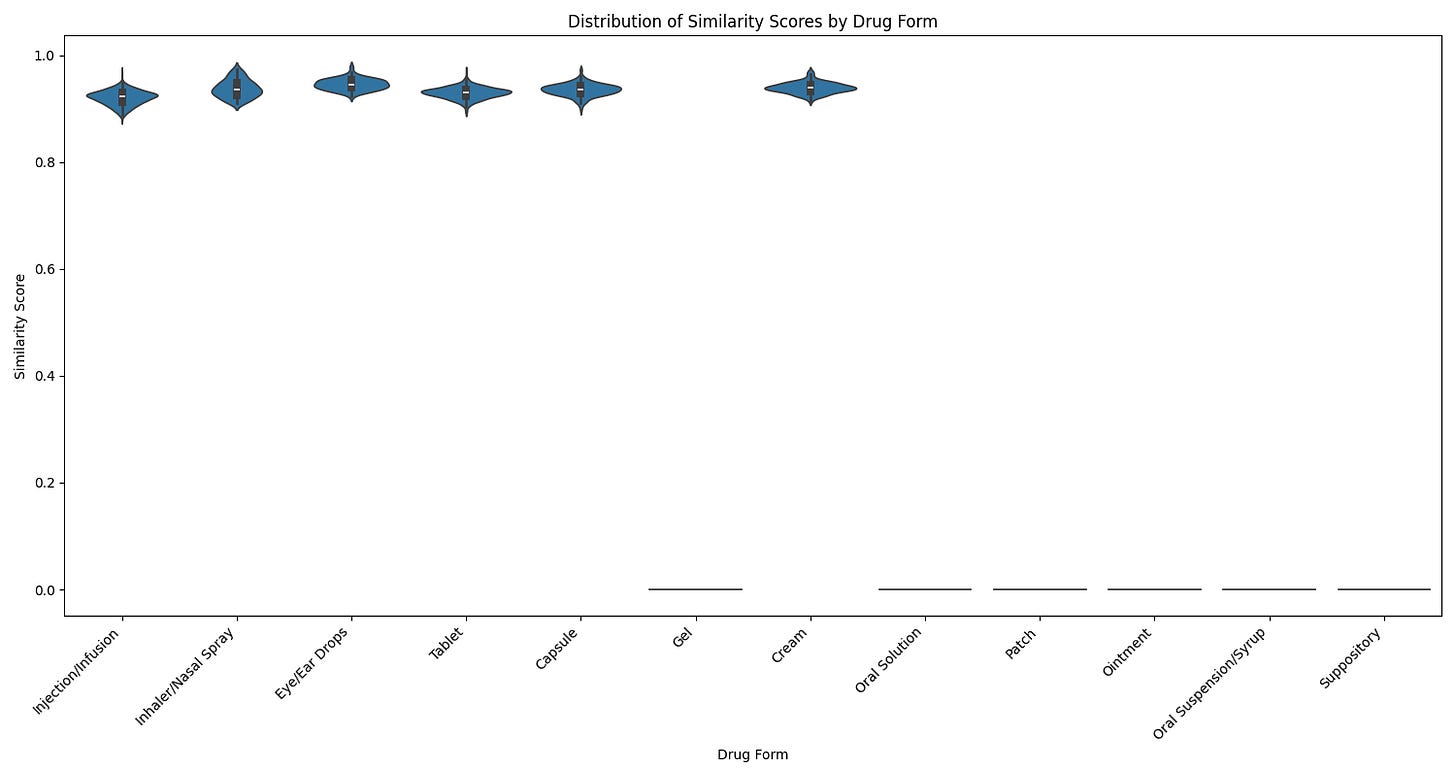
As you can see above, we had higher median scores (>0.95) for common forms and clear thresholds for reliable classifications.
The Confidence Threshold:
One of the most critical aspects of this classification system was determining when to trust the PubMedBERT decision. After analyzing thousands of classifications and their validation scores, a confidence threshold system had to be established that would help in making reliable decisions about the drug forms. So I identified three distinct confidence zones that help us balance accuracy with coverage:
The sweet spot emerged at 0.92—our high confidence threshold. When PubMedBERT returns a similarity score of ≥ 0.92, we're looking at classifications that are correct about 95% of the time. This covers approximately 75% of our drug database.
The story becomes more interesting in what I call the "medium confidence" zone"—scores between 0.85 and 0.91. These classifications are still quite reliable (around 85% accurate) but warrant a closer look. Often, these scores indicate drugs that legitimately come in multiple forms, like Amoxicillin, which is commonly available as both tablets and capsules.
Anything below 0.85 enters our low confidence zone. While these classifications might still be correct, the risk of error is too high for a medical application. Rather than make potentially incorrect assumptions, we mark these cases for manual review or additional verification.
Example: Amoxicillin
Let's look at how these thresholds work in practice with a common antibiotic, Amoxicillin:
Amoxicillin's Similarity Scores:
0.95 - Tablet ✅ High confidence (> 0.92)
0.87 - Capsule ⚠️ Medium confidence (0.85-0.91)
0.82 - Oral Solution ❌ Low confidence (< 0.85)
0.32 - Injection ❌ ❌ ❌ Very low confidence
The Power of Our Confidence Thresholds
For The High Confidence Classifications ≥ (0.92), 2,239 drugs (71.1% of our dataset) had a 93.0% accuracy rate. This means that when our system is highly confident, it's right 93% of the time, and this applies to over two-thirds of all drugs in our database. The remaining 157 drugs (7.0%), they disagreed on the form (Between GPT-4) and PubMedBERT Classification. For example, if we take 100 drugs from our high confidence group:
- For 93 of them, both models said the same thing (e.g., both said "Tablet" for Amoxicillin)
- For 7 of them, they disagreed (e.g., GPT-4 said "Tablet" but PubMedBERT suggested "Capsule")
So When they agree, we have higher confidence that the classification is correct because two different AI models came to the same conclusion independently.
For The Medium Confidence Classifications (0.85-0.91), 739 drugs (23.5% of our dataset) had a 67.5% accuracy rate. These cases often represent drugs that legitimately come in multiple forms. For the Low Confidence Classifications (<0.85), 72 drugs (5.5% of our dataset) had a very low accuracy rate.
This small group of drugs typically requires manual verification or represents complex cases with multiple possible forms.
Hope for a better streamlined Flow
Our dual-AI classification system achieved several key milestones:
Like I stated earlier from the beginning of this write up, the main aim of creating this was to help streamline our process, in hopes to make the flow and personalization result better . I will also be open-sourcing this Data Set on Hugging Face and the analysis code will be on my Github.
Coverage and Accuracy
- Successfully classified 3,150 drugs across 12 standardized forms
- Achieved 93% accuracy for high-confidence classifications
- Identified alternative forms for drugs with multiple valid formulations
Up next, I will be cleaning up the files and then work on pushing them to my repo. I have made the UI Design update on Figma as you can see from the beginning, I now have to go figure how to implement this with Cursor :)
I do hope you find the dataset useful in your own analysis work, if you do let me know and don’t forget to drop your suggestions and feedback below. That’s 10 minutes see you soon.